グラフつくるとことは禁断探索法と一緒。
def evaluate(nodes, adj, sol, alpha):
s = [0 for i in nodes]
d = [0 for i in nodes]
bal = 0
for i in nodes:
if sol[i] == 0:
bal += 1
else:
bal -= 1
for j in adj[i]:
if sol[j] == sol[i]:
s[i] += 1
else:
s[i] -= 1
cost = 0
for i in nodes: cost += d[i]
cost /= 2
cost += alpha*abs(bal)
return cost, bal, s, d
def find_move_rnd(n, sol, alpha, s, d, bal):
istar = random.randint(0,n-1)
part = sol[istar]
if part == 0 and bal > 0 or part == 1 and bal < 0:
penalty = -2*alpha
else:
penalty = 2*alpha
delta = s[istar] - d[istar] + penalty
return istar, delta
def update_move(adj, sol, s, d, bal, istar):
part = sol[istar]
sol[istar] = 1-part
s[istar], d[istar] = d[istar], s[istar]
for j in adj[istar]:
if sol[j] == part:
s[j] -= 1
d[j] += 1
else:
s[j] += 1
d[j] -= 1
if part == 0:
bal -= 1
else:
bal += 1
return bal
def estimate_temperature(n, sol, s, d, bal, initprob):
ntrials = 10 * len(sol)
nsucc = 0
deltaZ = 0.0
for i in range(0,ntrials):
part = random.randint(0,1)
istar, delta = find_move_rnd(n, sol, alpha, s, d, bal)
if delta > 0:
nsucc += 1
deltaZ += delta
if nsucc != 0:
deltaZ /= nsucc
T = -deltaZ/math.log(initprob)
return T
def metropolis(T, delta):
if delta <= 0 or random.random() < math.exp(-(delta)/T):
return True
else:
return False
def annealing(nodes, adj, sol, initprob, tempfactor, sizefactor, cutoff, freezelim, minpercent, alpha):
n = len(nodes)
z,bal,s,d = evaluate(nodes, adj, sol, alpha)
if bal == 0:
solstar, zstar = list(sol), z
T = estimate_temperature(n, sol, s, d, bal, initprob)
nfrozen = 0
part = 0
while nfrozen < freezelim:
changes, trials = 0,0
while trials < n*sizefactor and changes < n*cutoff:
trials += 1
istar, delta = find_move_rnd(n, sol, alpha, s, d, bal)
if metropolis(T, delta):
changes += 1
bal = update_move(adj, sol, s, d, bal, istar)
z += delta
if bal == 0:
if z < zstar and part == 0:
solstar, zstar = list(sol), z
nfrozen = 0
T *= tempfactor
if float(changes)/trials < minpercent:
nfrozen += 1
return solstar, zstar
if __name__ == "__main__":
import networkx as nx
import matplotlib.pyplot as plt
num_nodes = 50
nodes, adj = rnd_graph_fast(num_nodes,0.3)
G=nx.Graph()
for i in range(num_nodes):
G.add_node(i)
for i in range(num_nodes-1):
for j in adj[i]:
if i < j:
G.add_edge(i,j)
pos=nx.spring_layout(G)
initprob = .4 # initial acceptance probability
sizefactor = 16 # for det. # tentatives at each temp.
tempfactor = .95 # cooling ratio
freezelim = 5 # max number of iterations with less that minpercent acceptances
minpercent = .02 # fraction of accepted moves for being not frozen
alpha = .05 # imballance factor
N = len(nodes) # neighborhood size
L = N*sizefactor # number of tentatives at current temperature
sol = construct(nodes)
z,bal,s,d = evaluate(nodes, adj, sol, alpha)
sol,z = annealing(nodes, adj, sol, initprob, tempfactor, N, L, freezelim, minpercent, alpha)
zp,bal,sp,dp = evaluate(nodes, adj, sol, alpha)
rnodelist = []
bnodelist = []
for i in range(len(sol)):
if sol[i] == 1:
rnodelist.append(i)
else:
bnodelist.append(i)
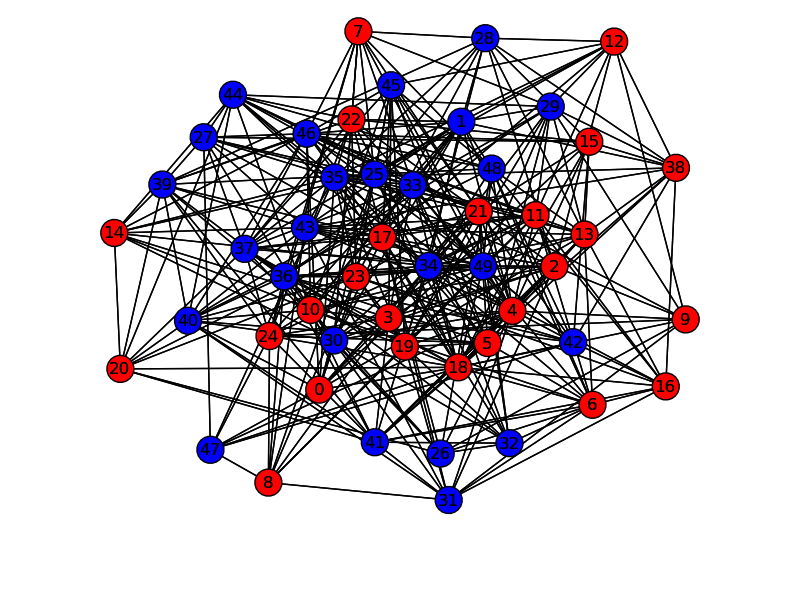
nx.draw(G,pos,nodelist=rnodelist,node_color='r')
nx.draw(G,pos,nodelist=bnodelist,node_color='b')
plt.savefig("path.png")

 パターン認識と機械学習 下 - ベイズ理論による統計的予測
パターン認識と機械学習 下 - ベイズ理論による統計的予測
 パターン認識と機械学習 上 - ベイズ理論による統計的予測
パターン認識と機械学習 上 - ベイズ理論による統計的予測 Programming the Semantic Web
Programming the Semantic Web
 ファーマコキネティクス―演習による理解
ファーマコキネティクス―演習による理解 メタヒューリスティクスの数理
メタヒューリスティクスの数理