kNN, Naibe Bayesm, SVM, (Random Forrest)をScikit-learnでやってみた。データはiPython NotebookでReSTで出力したものをpandocでmarkdown_strictに変換しなおしてblogに貼り付けた。
描画用のヘルパー関数とデータセットの生成
from matplotlib.colors import ListedColormap
import Image
from numpy import *
from pylab import *
import pickle
def myplot_2D_boundary(X,y):
x_min, x_max = X[:, 0].min() - 1, X[:, 0].max() + 1
y_min, y_max = X[:, 1].min() - 1, X[:, 1].max() + 1
xx, yy = np.meshgrid(np.arange(x_min, x_max, 0.02),
np.arange(y_min, y_max, 0.02))
Z = clf.predict(np.c_[xx.ravel(), yy.ravel()])
Z = Z.reshape(xx.shape)
plt.figure()
plt.pcolormesh(xx, yy, Z, cmap=cmap_light)
plt.scatter(X[:, 0], X[:, 1], c=y, cmap=cmap_bold)
plt.xlim(xx.min(), xx.max())
plt.ylim(yy.min(), yy.max())
plt.show()
with open('points_normal.pkl', 'r') as f:
class_1 = pickle.load(f)
class_2 = pickle.load(f)
labels = pickle.load(f)
X_normal = np.r_[class_1, class_2]
y_normal = labels
with open('points_ring.pkl', 'r') as f:
class_1 = pickle.load(f)
class_2 = pickle.load(f)
labels = pickle.load(f)
X_ring = np.r_[class_1, class_2]
y_ring = labels
with open('points_normal_test.pkl', 'r') as f:
class_1 = pickle.load(f)
class_2 = pickle.load(f)
labels = pickle.load(f)
X_normal_test = np.r_[class_1, class_2]
y_normal_test = labels
with open('points_ring_test.pkl', 'r') as f:
class_1 = pickle.load(f)
class_2 = pickle.load(f)
labels = pickle.load(f)
X_ring_test = np.r_[class_1, class_2]
y_ring_test = labels
cmap_light = ListedColormap(['#FFAAAA', '#AAFFAA'])
cmap_bold = ListedColormap(['#FF0000', '#00FF00'])
kNN
import numpy as np
import matplotlib.pyplot as plt
from matplotlib.colors import ListedColormap
from sklearn import neighbors, datasets
clf = neighbors.KNeighborsClassifier(3)
clf.fit(X_normal, y_normal)
myplot_2D_boundary(X_normal,y_normal)
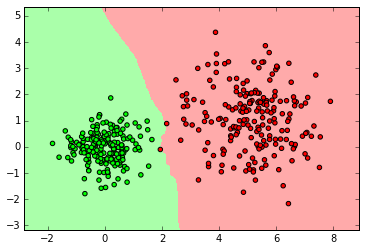
clf = neighbors.KNeighborsClassifier(3)
clf.fit(X_ring, y_ring)
myplot_2D_boundary(X_ring,y_ring)
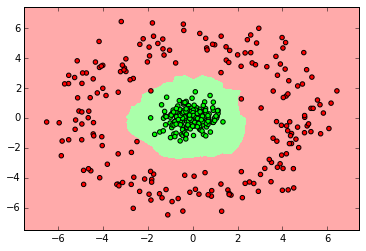
ベイズ
from sklearn.naive_bayes import GaussianNB
clf = GaussianNB()
clf.fit(X_normal, y_normal)
labels_pred = clf.predict(X_normal_test)
print "Number of mislabeled points out of a total %d points : %d" % (y_normal_test.shape[0],(y_normal_test != labels_pred).sum())
myplot_2D_boundary(X_normal,y_normal)
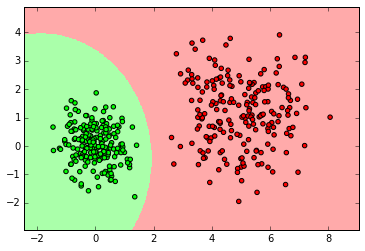
clf = GaussianNB()
clf.fit(X_ring, y_ring)
labels_pred = clf.predict(X_ring_test)
print "Number of mislabeled points out of a total %d points : %d" % (y_ring_test.shape[0],(y_ring_test != labels_pred).sum())
myplot_2D_boundary(X_ring,y_ring)
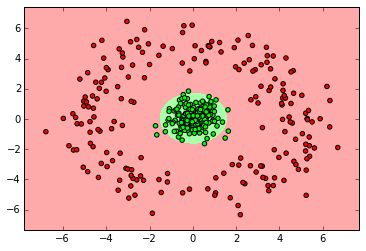
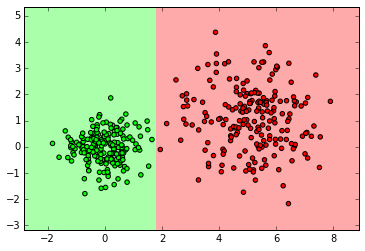
SVM
from sklearn import svm
clf = svm.SVC()
clf.fit(X_normal, y_normal)
labels_pred = clf.predict(X_normal_test)
print "Number of mislabeled points out of a total %d points : %d" % (y_normal_test.shape[0],(y_normal_test != labels_pred).sum())
myplot_2D_boundary(X_normal, y_normal)
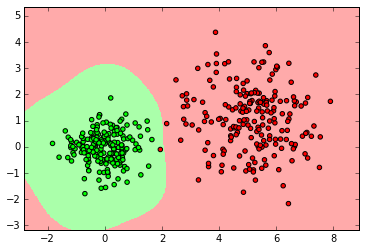
clf = svm.SVC()
clf.fit(X_ring, y_ring)
labels_pred = clf.predict(X_ring_test)
print "Number of mislabeled points out of a total %d points : %d" % (y_ring_test.shape[0],(y_ring_test != labels_pred).sum())
myplot_2D_boundary(X_ring,y_ring)
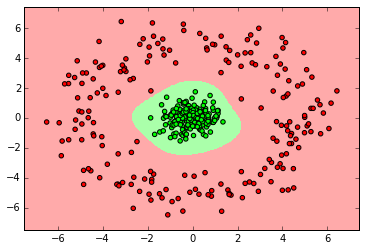
Random Forest
from sklearn.ensemble import RandomForestClassifier
clf = RandomForestClassifier(n_estimators=10)
clf.fit(X_normal, y_normal)
labels_pred = clf.predict(X_normal_test)
print "Number of mislabeled points out of a total %d points : %d" % (y_normal_test.shape[0],(y_normal_test != labels_pred).sum())
myplot_2D_boundary(X_normal,y_normal)
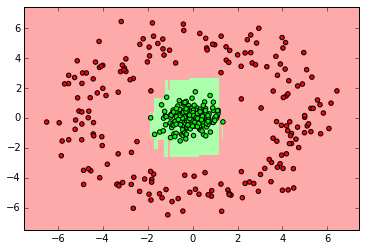
from sklearn.ensemble import RandomForestClassifier
clf = RandomForestClassifier(n_estimators=10)
clf.fit(X_ring, y_ring)
labels_pred = clf.predict(X_ring_test)
print "Number of mislabeled points out of a total %d points : %d" % (y_ring_test.shape[0],(y_ring_test != labels_pred).sum())
myplot_2D_boundary(X_ring,y_ring)




 玉蘭 (文春文庫)
玉蘭 (文春文庫) IPython Interactive Computing and Visualization Cookbook
IPython Interactive Computing and Visualization Cookbook ダイレクト向かい飛車徹底ガイド (マイナビ将棋BOOKS)
ダイレクト向かい飛車徹底ガイド (マイナビ将棋BOOKS) 相振り飛車を指しこなす本〈1〉 (最強将棋21)
相振り飛車を指しこなす本〈1〉 (最強将棋21)










 ハヅキさんのこと (講談社文庫)
ハヅキさんのこと (講談社文庫)