12122018 chemoinformatics rdkit
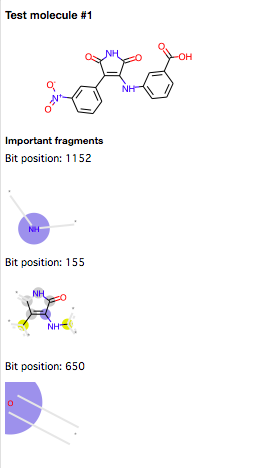
Interpretaion of the QSAR model with fingerprints is difficult because its features don't show substructures but position of bits. The latest version of RDKIt has implemented the new fingerprint bit rendering code, so we are easily able to understand the meaning of the bit as a substructure (fragment)
Now, I wrote some code to visualize important fragments of the compounds and uploaded it to GitHub.
If you are familiar with Kinase inhibitors, I am sure that you will notice its usefulness.